splitify¶
splitify writes out differences of masses for the splits of the tree.
usage: splitify [options] placefile(s)
Options¶
| -o | Specify the filename to write to. |
| --out-dir | Specify the directory to write files to. |
| --prefix | Specify a string to be prepended to filenames. |
| --point-mass | Treat every pquery as a point mass concentrated on the highest-weight placement. |
| --pp | Use posterior probability for the weight. |
| --kappa | Specify the exponent for scaling between weighted and unweighted splitification. default: 1 |
| --rep-edges | Cluster neighboring edges that have splitified euclidean distance less than the argument. |
| --epsilon | The epsilon to use to determine if a split matrix’s column is constant for filtering. default: no filtering |
Details¶
The first step to perform edge PCA is to make a matrix with rows indexed by the samples, and columns by the edges of the tree.
The 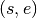 entry of this matrix is the difference between the distribution of mass on either side of edge
entry of this matrix is the difference between the distribution of mass on either side of edge  for the sample
for the sample  .
Specifically, it is the amount of mass on the distal (non-root) side of edge
.
Specifically, it is the amount of mass on the distal (non-root) side of edge  minus the amount of mass on the proximal side.
The matrix is indexed such that the first numerical column is edge labeled 0 in the reference tree.
The splitify subcommand simply writes out this matrix.
minus the amount of mass on the proximal side.
The matrix is indexed such that the first numerical column is edge labeled 0 in the reference tree.
The splitify subcommand simply writes out this matrix.
Specifying --rep-edges x will only take representatives from collections of neighboring edges whose Euclidean distance between splitified columns is less than x.
The --kappa option enables a componentwise transformation 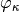 on the entries of this matrix.
on the entries of this matrix.
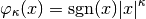
where the  parameter can any non-negative number.
This parameter scales between ignoring abundance information (
parameter can any non-negative number.
This parameter scales between ignoring abundance information (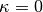 ), using it linearly (
), using it linearly (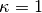 ), and emphasizing it (
), and emphasizing it ( ).
).