Sequence alignment from the
phylogenetic
perspective
Erick Matsen
In phylogenetics, homology means..
Quiz: Why?
Structural alignment is another perspective
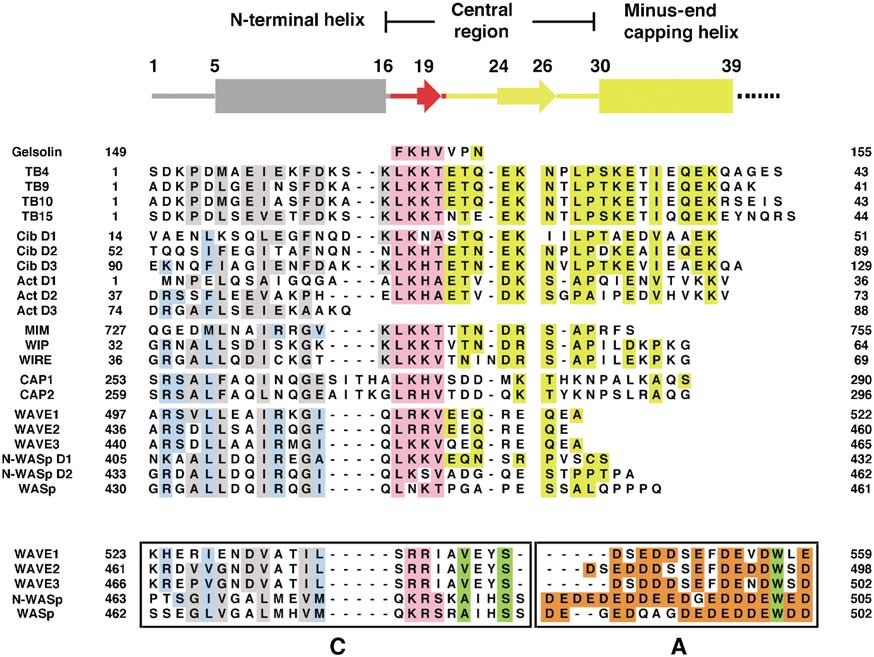
Sequence alignment for phylogenetics is hard!
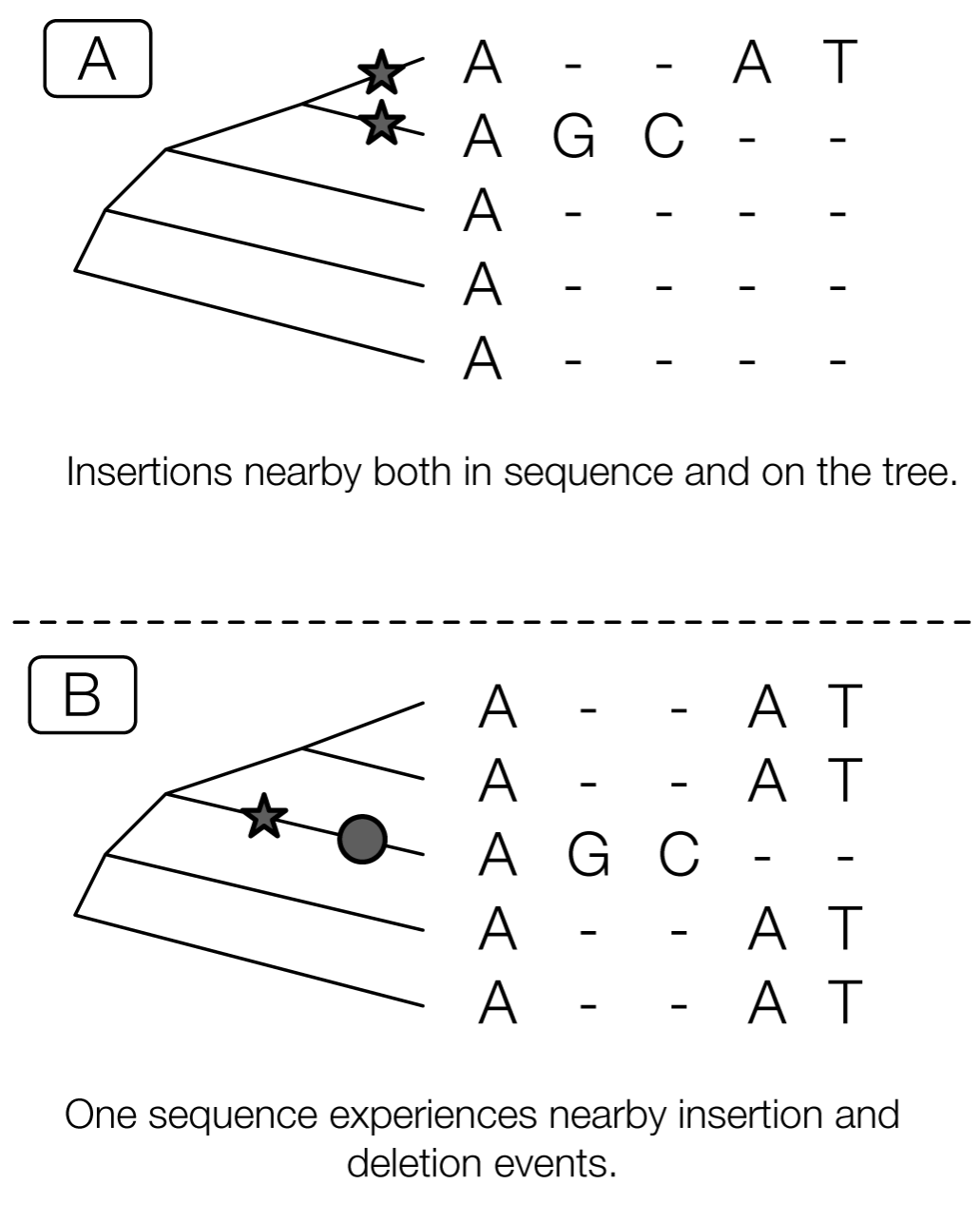
Sometimes it’s more or less impossible
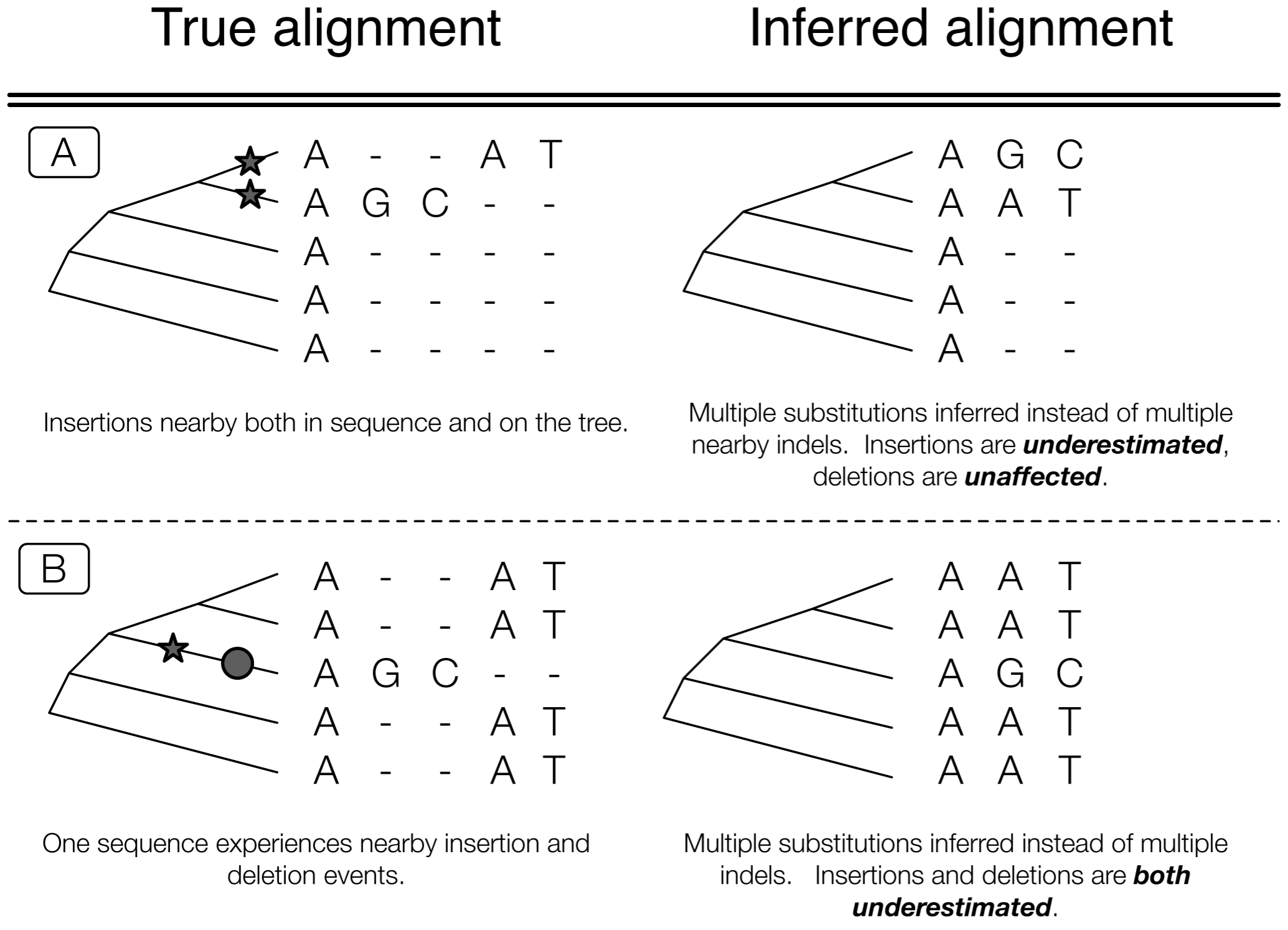
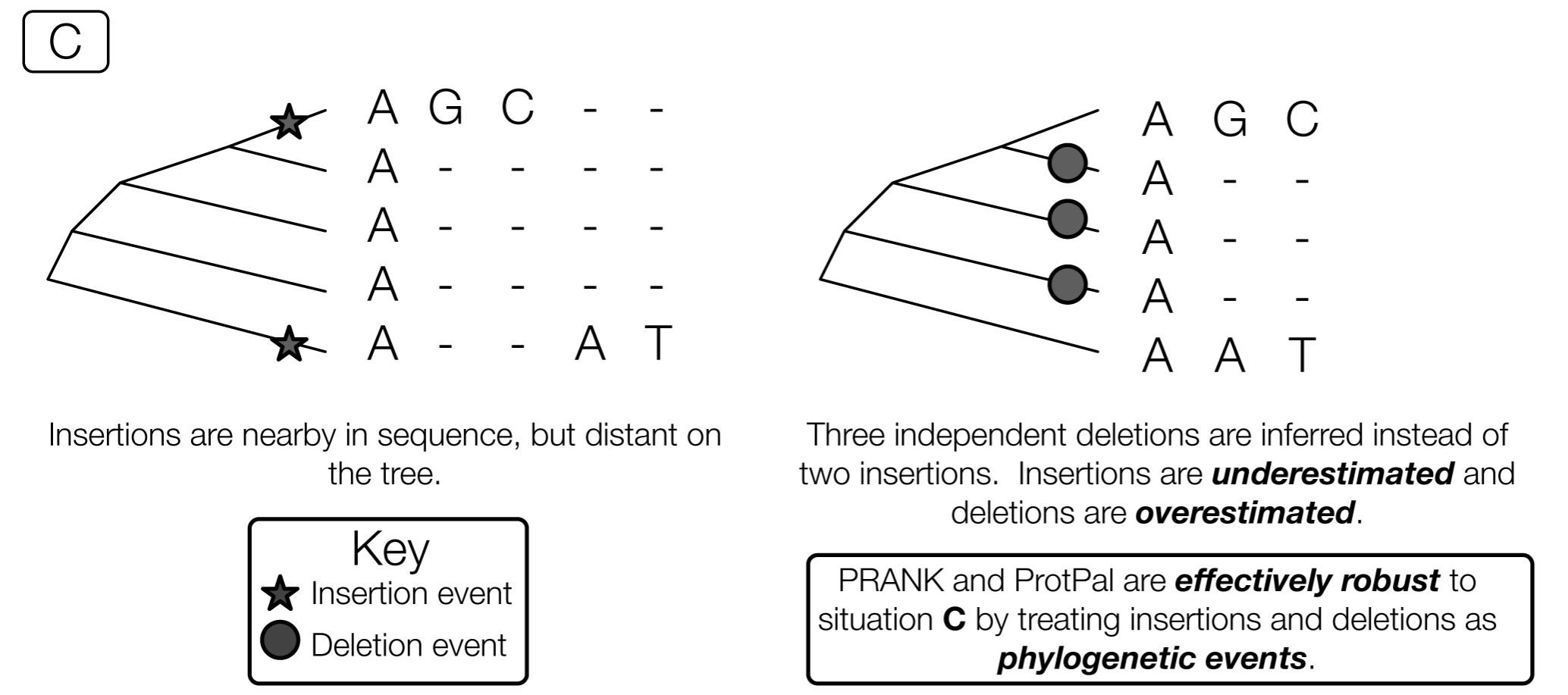
Phylogenetic approach is best
Clustal W vs PRANK
PRANK is a smart “hack”
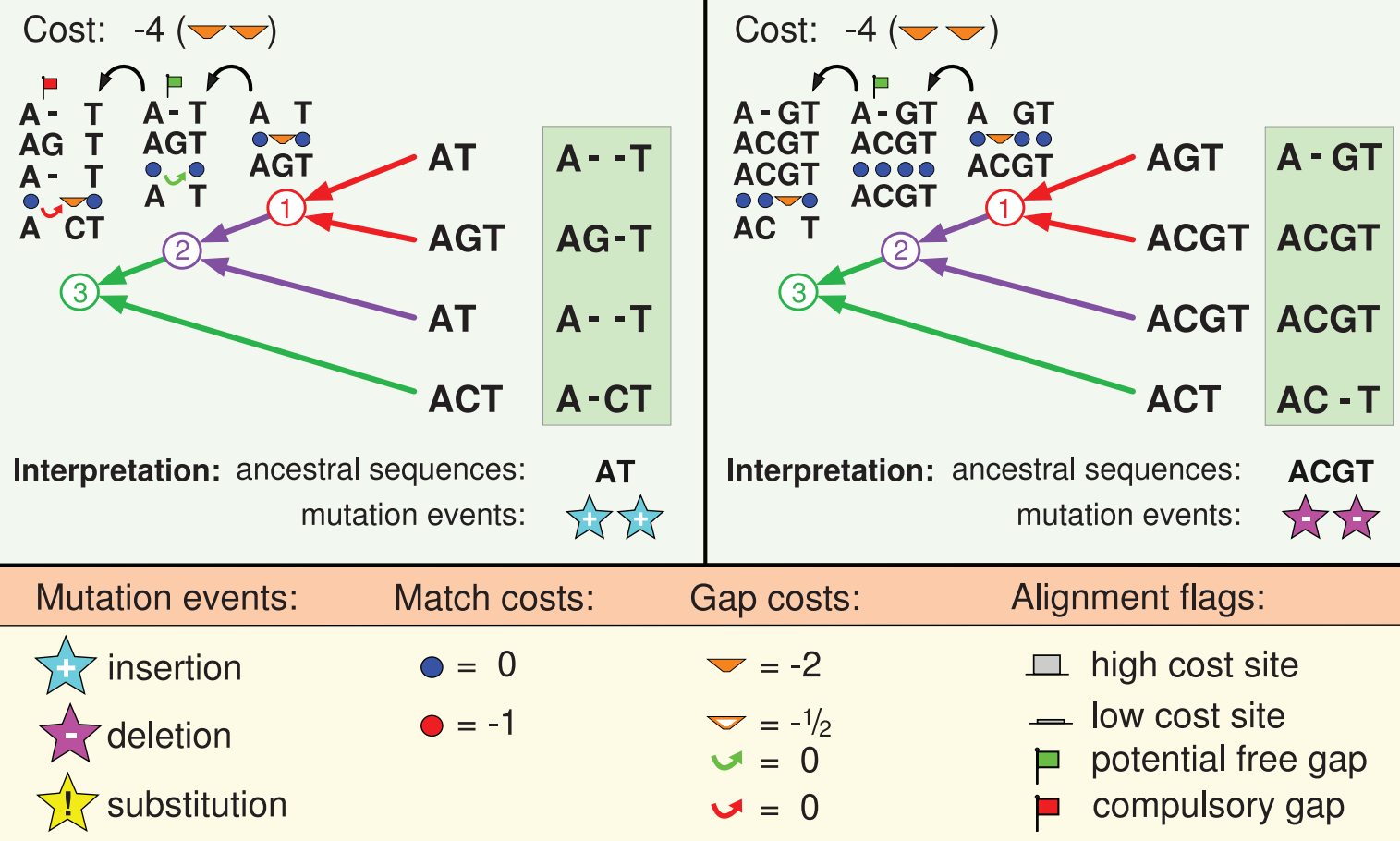
BAli-Phy is fully model-based
BAli-Phy is fully model-based
BAli-Phy is fully model-based
BAli-Phy is fully model-based
BAli-Phy is fully model-based
Note how BAli-Phy can trace the history of each site through
the
tree, and the columns are simply a reflection of that.
BAli-Phy estimates alignment uncertainty
BAli-Phy is fully Bayesian, so it has a rigorous notion of alignment uncertainty, which can be elegantly summarized by that software:

BAli-Phy summary
- Co-estimate tree & alignment using Bayesian methods
- Can also use codon models for selection analysis; integrating out alignments eliminates artifacts
- Very computationally expensive: 60 sequences may take days
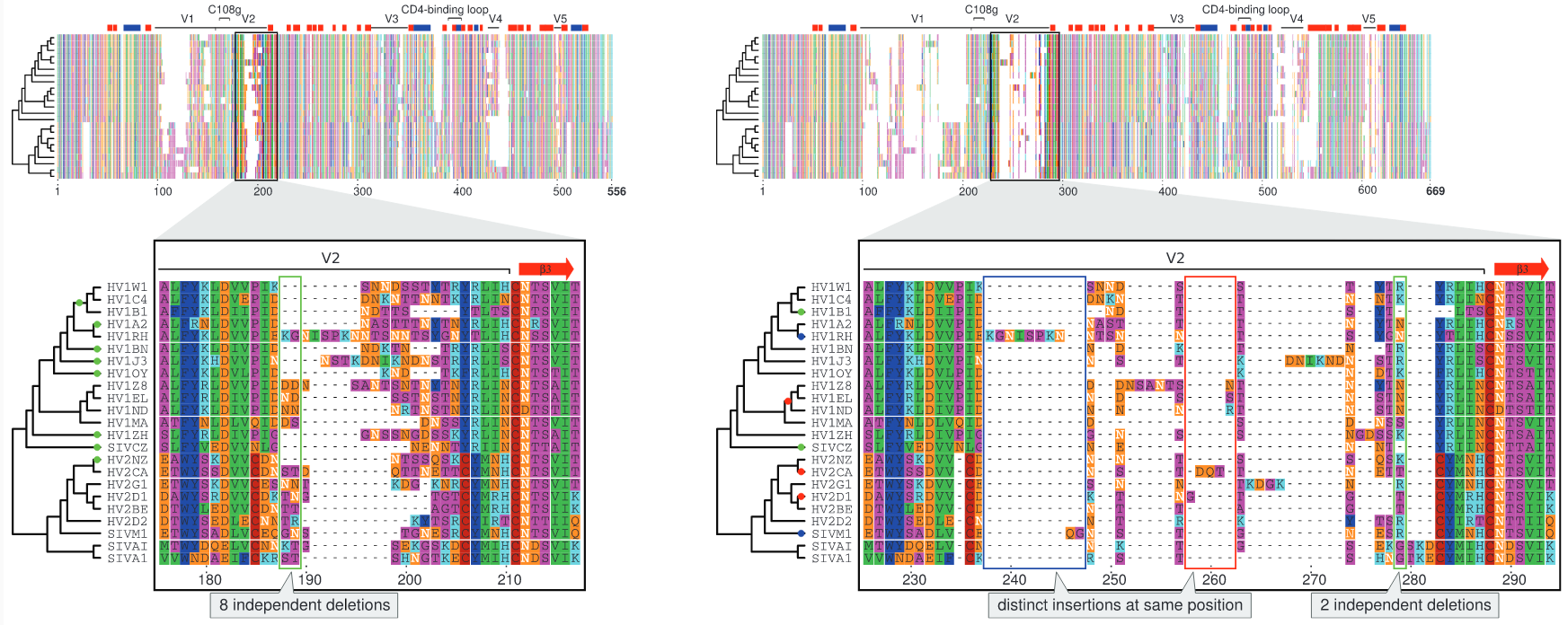
What’s funny about this protein-coding alignment?
What’s funny about this protein-coding alignment?
Multiple sequence alignment summary
- If you really care about alignments (and trees) use BAli-Phy
- MAFFT ranked the best in a recent benchmark by the authors of PRANK concerning ancestral sequence reconstruction
-
If you want good alignment using PRANK
with a lovely web-based GUI, use Wasabi -
If you want a quick and dirty alignment, use Muscle
(esp. if your sequences aren’t too divergent) - Do not use any variant of Clustal, even if your friends say to.
- Use a codon alignment if you have protein-coding genes.